

In this blog i will take you through processing fMRI data and make your fMRI data ready to be used in your analysis. We will use FSL’s FEAT tool for achieving the required results. Take a look at the below video to know how we obtain MRI scans.
There are mainly 2 types of fMRI, task fMRI and Resting state fMRI, the first open corresponds to when the subject is performing some task like breathing, tapping fingers inside the scanner and the latter corresponds to the subject being still and trying not to fall asleep inside the scanner. Both of them have a very different set of analysis to perform and we’ll go through preprocessing both of the fMRI data types. fMRI is intrinsically more noisier so we will need more tools for the preprocessing

Click the link to reach my tutorial on processing structural MRI
For this blog we will use the fMRI data of a CN (cognitively normal) subject from ADNI dataset, you can access the data , here : github link ,
I have changed the file structure of the data for ease of programming
The data that ADNI provides is in .dcm format and FEAT working only with 4D NIFTI (3D+time) and it’s recommended that the images be brain extracted.
Utilize tools like dcm2niix for conversion of .dcm to .nifti

you can see there are non brain material in the above gif for better analysis we would like them to be removed,, if we use bet tool on a 4D image then it will take only the first 3D MRI image and return only one brain extracted MRI image.
So we break the fMRI data into structural MRI image for each point in the timestamp and then perform brain extraction and then again put them together to achieve brain
#Extract individual volumes (timepoints) from the 4D dataset
fslsplit 002_S_4262_1.nii volumes -t
#Perform brain extraction on each volume separately
for volume in volumes*.nii.gz; do
base_name=$(basename "$volume" .nii.gz)
bet "$volume" "brain_extracted_$base_name"
done
#Merge the brain-extracted volumes back into a 4D dataset
fslmerge -t out_brain_extracted.nii.gz brain_extracted_*.nii.gz
#delete all the intermediate files
rm -r brain_extracted*.nii.gz volumes*.nii.gz
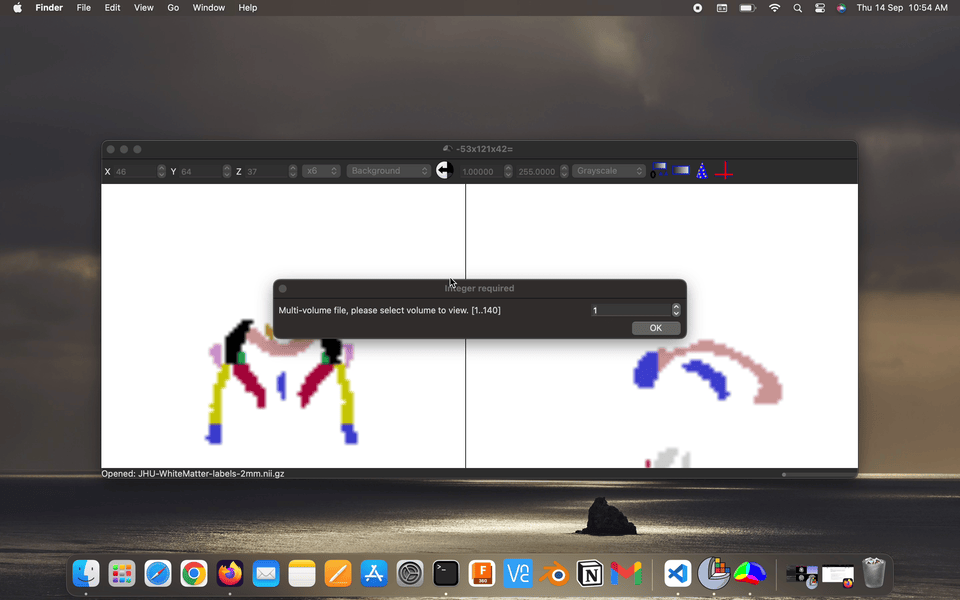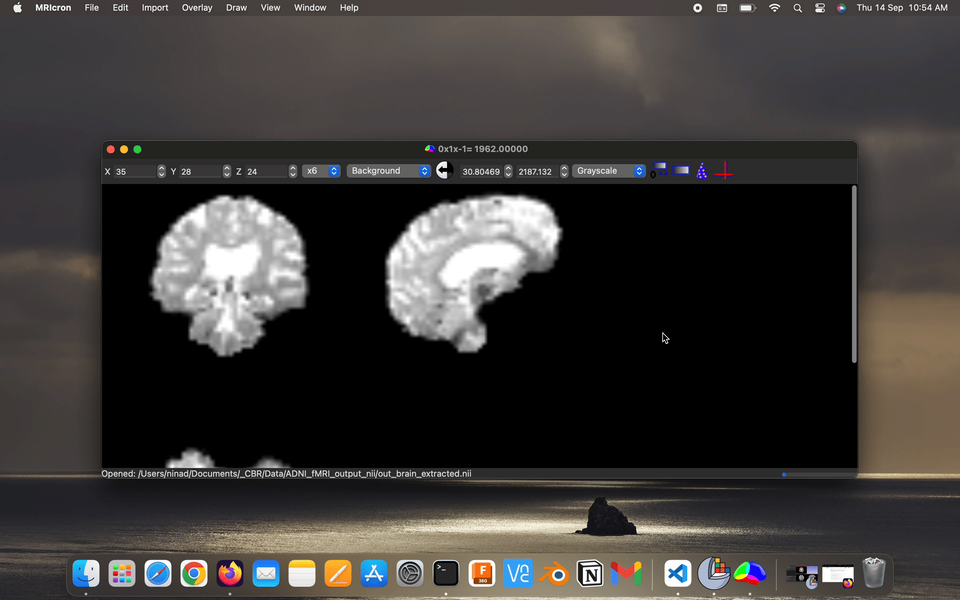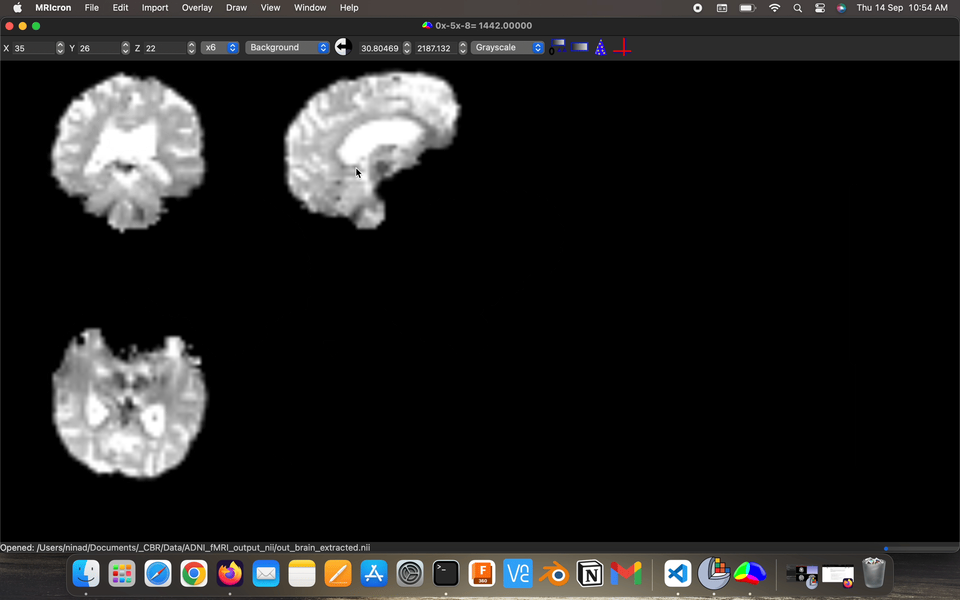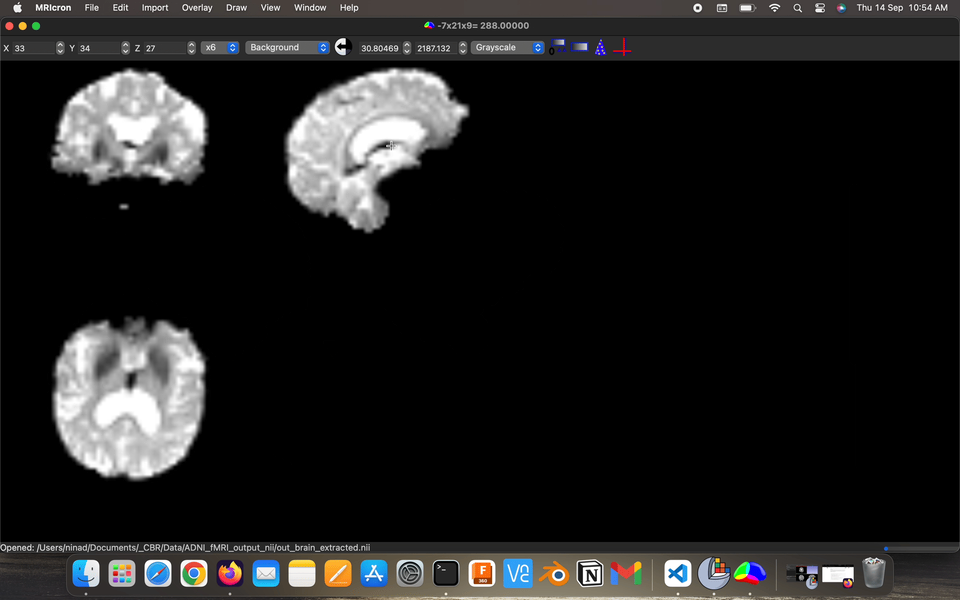
Fact: the energy consumption of the various parts of the brains, brains has ~2% body weight but ~20% energy consumption.


In task fMRI analysis, we know what to expect / look for ,, ie. there will be some noticeable changes in intensity and we model that using probability distributions or Gaussian linear models, but in Resting State fMRI we don’t know what to expect and there adds noise in every step of the process, and these noise can dominate in the signal we are interested in thus dominate our final results

Using FSLeyes visualize the high-res anatomical T1w images and it’s corresponding fMRI image
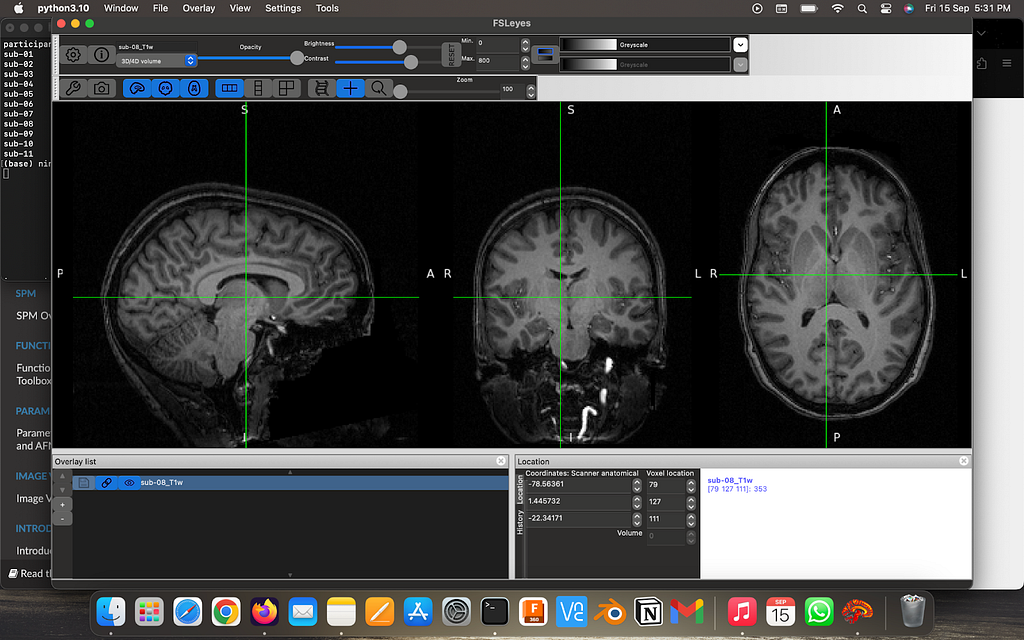
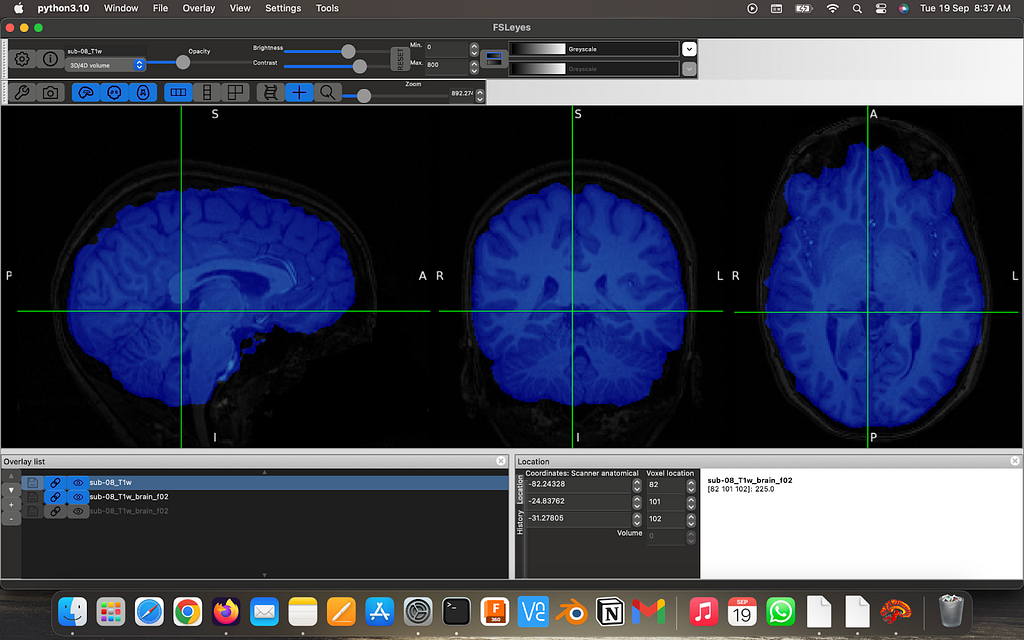
Perform brain extraction on the high-res image
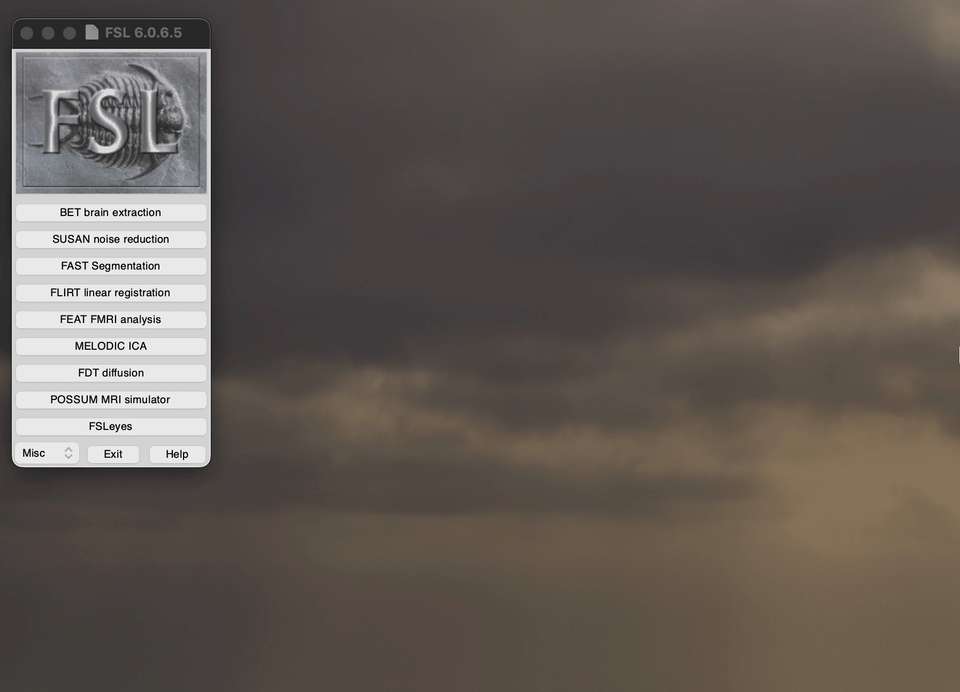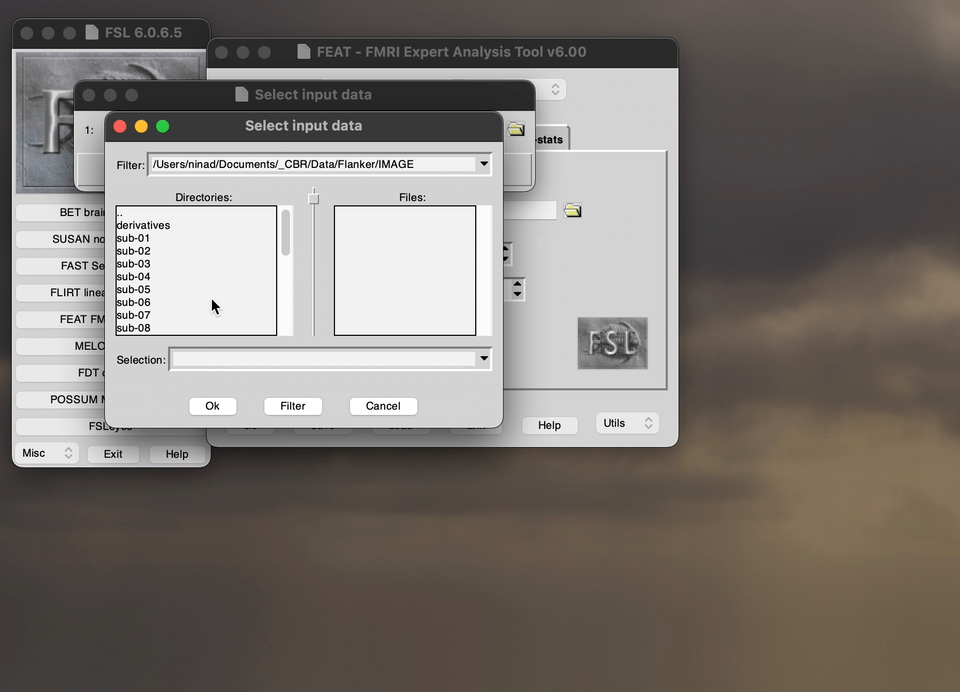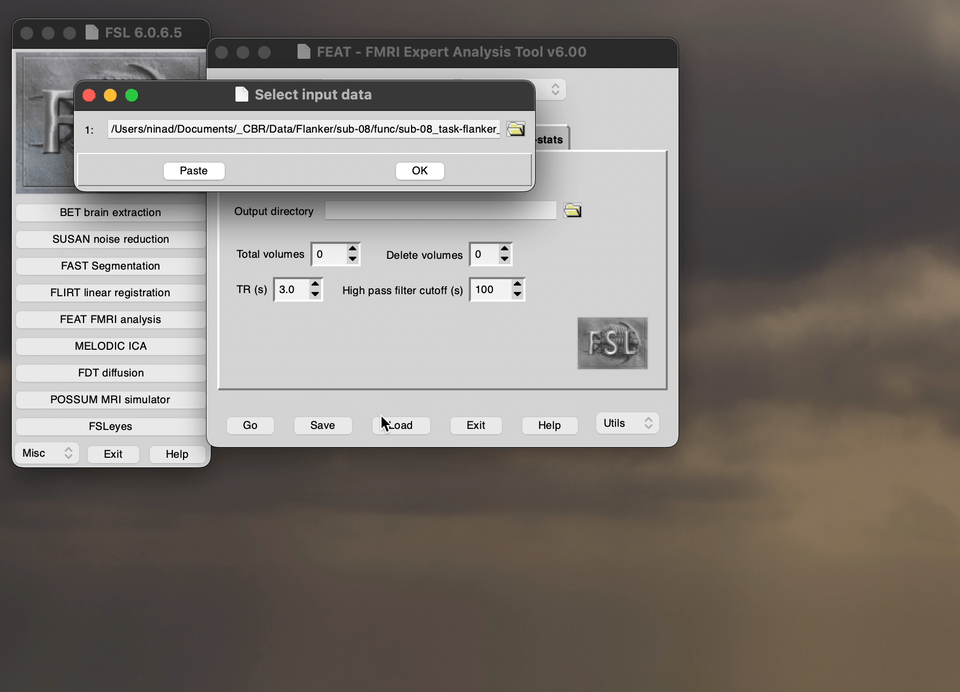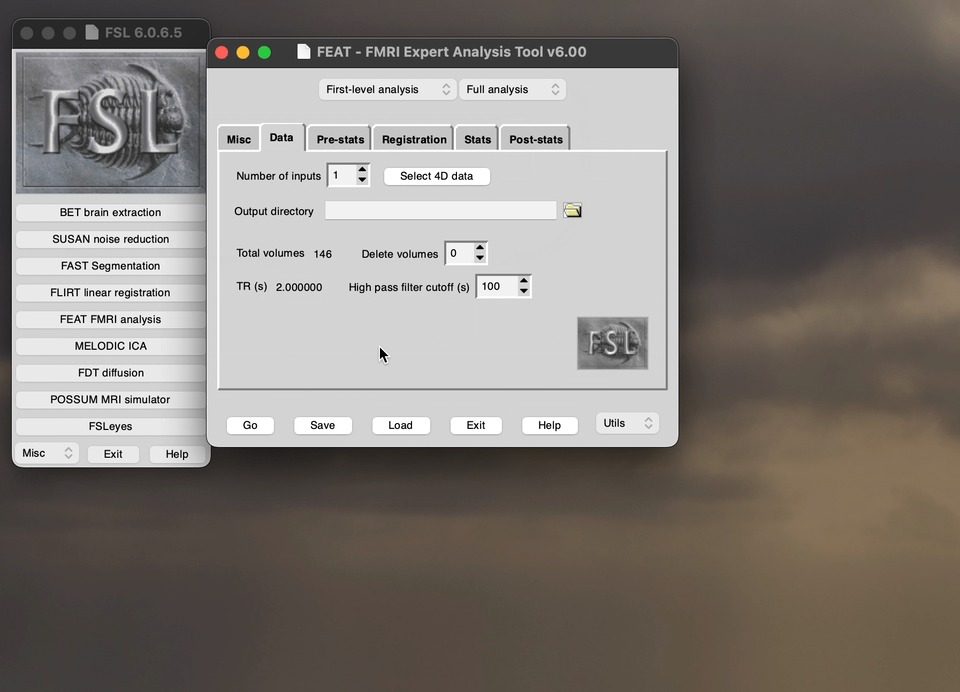
The FEATtool is used to preprocess the fMRI, the below demo directs you what to choose the options as before you run the preprocessing. Specifically, this demo shows you how to upload the data into the preprocessing setup tool.
Snippet of the Pre-stats tab in the FEAT toolbox

Demo on what slice time correction means and how it affects the analysis.

Options to select on the registration tab

Click Go and you will start the analysis,
Below is the summary of the analysis performed by the FEAT tool

Visualize the output preprocessed fMRI data saved as filtered_func_data.nii.gz
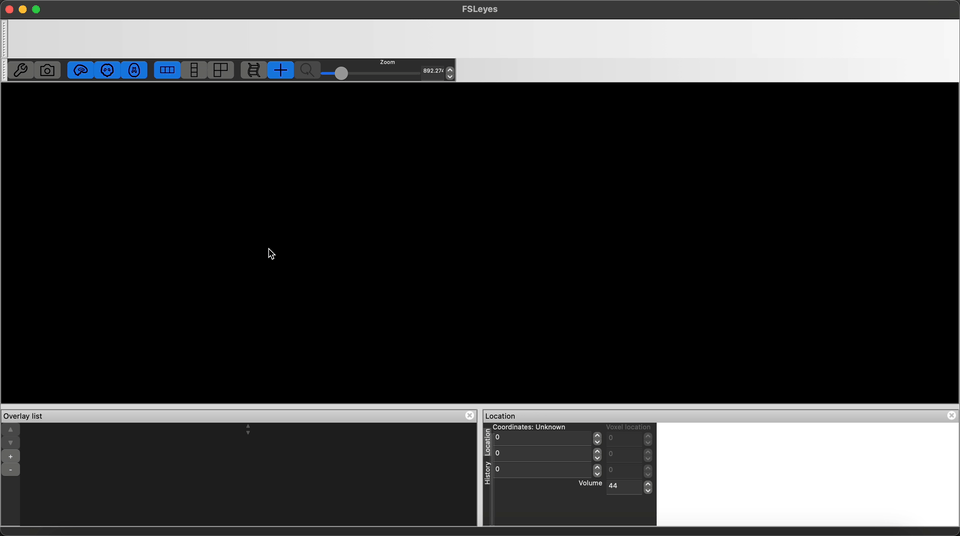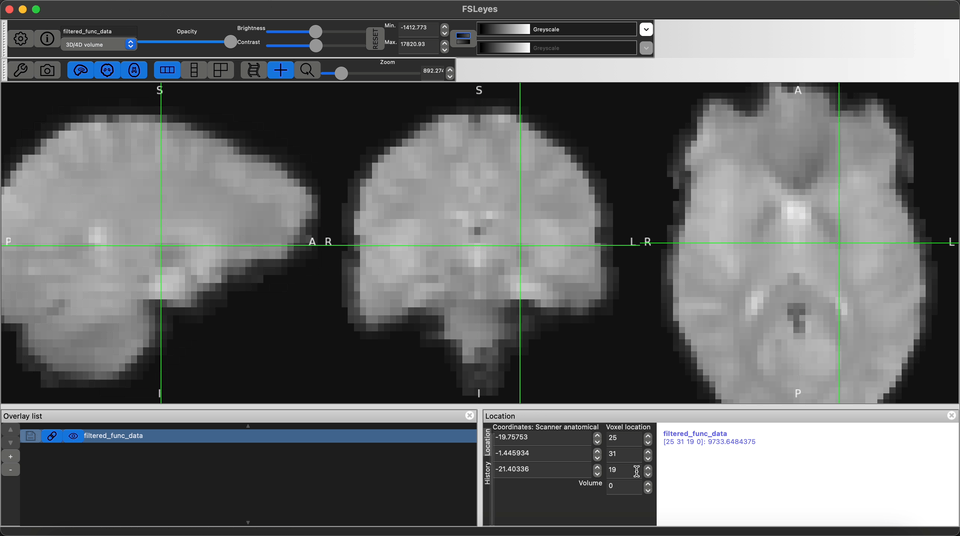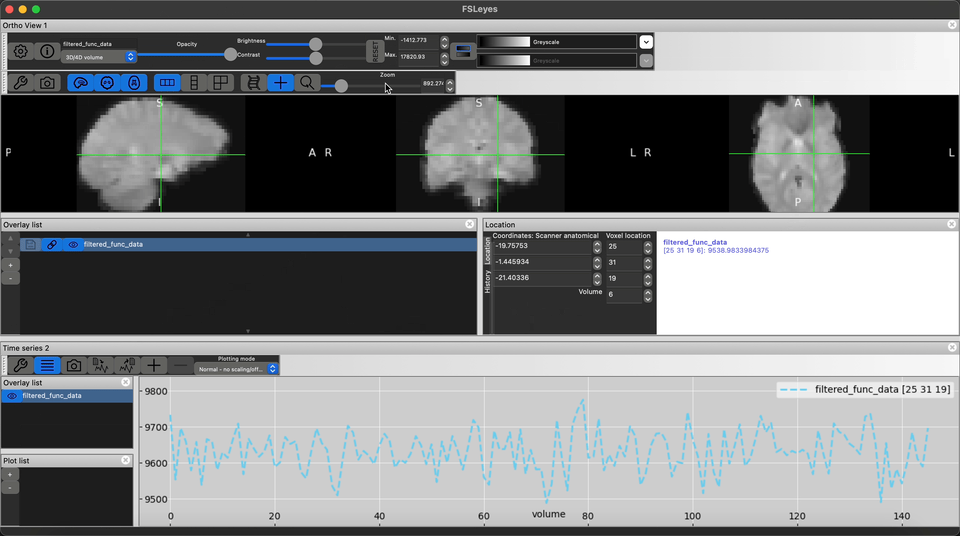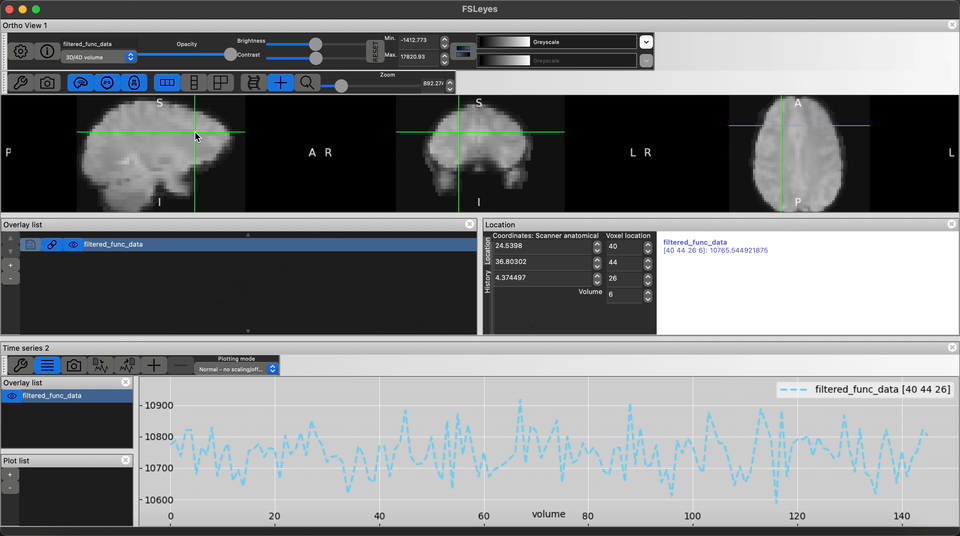
Refrences