
This tutorial should help you to work your way through FSL
For this article / tutorial i have used the a MRI image from OASIS dataset which you can access using this link 🔗

Extract information about the MRI image
fslinfo mri_image.nii.gz
the output of the above command gives a lot of information about the MRI image file and not about the subject
dim1 128 dim2 256 dim3 256 shows that this is a 128x256x256 matrix
the output also mentions that each voxel is 1.25 mm x 1 mm x 1 mm
FSL assumes that the brain is made up of small volumes and this can be visualized below
we are going remove the skull non tissue part of the brain
bet2 mri_image.nii.gz skull_stripped -f 0.5

bet2 has a important parameter Fractional Intensity Parameter (f) which is set to a default value of 0.5, smaller values of f give larger brain outline estimates.
Assignment : Try using various different values of F and see what happens
Not all the part of the MRI image is important, we can discard the lower head and neck part of the MRI image so that we can focus more on the brain, we use a tool roboustfov
robustfov -v -i skull_stripped.nii.gz -r roiimage
Final FOV is:
0.000000 128.000000 0.000000 256.000000 85.000000 170.000000
Xmin Xmax Ymin Ymax Zmin Zmax
fslinfo roiimage.nii.gz
MRI image is now of the shape 128x256x170
Sometimes the MRI images are not in the standard orientation like they can be flipped, mirrored , then we can run fslreorient2std this is not a registration tool and can perform only 90°, 180°, 270° rotations
fslreorient2std roiimage.nii.gz
#to create a new output file with the orienattion of the standard MNI template
fslreorient2std roiimage.nii.gz reoriented
1 0 0 0
0 1 0 0
0 0 1 0
0 0 0 1
the output is a identity matrix, implying that the MRI image in the correct orientation ie,
now we have a skull stripped brain image and we will segment the brain into various parts, normally White matter, Grey Matter & Cerebrospinal fluid (CSF) in case of T1 weighted MRI images, if there are big lesions in the brain , we can classify them into another class
we use FAST tool to achieve brain and along with segmentation it also corrects the bias in the the MRI images. FAST uses Mori-Tanaka approach to classify the voxel into a class and each voxel can belong to different classes, the probability of it belonging to a class is showed in it’s intensity
Assignment: Find out what is Bias in the MRI images, Is it Spatially invariant or not.
HINT : check -b option in fast
FAST is a iterative tool and there is a trade off between number of iterations (time taken) and the accuracy of the segmentation.
fast -o fast_out -b -B -t 1 --iter=10 -v roiimage.nii.gz
output :
.
├── fast_out_bias.nii.gz
├── fast_out_mixeltype.nii.gz
├── fast_out_pve_0.nii.gz
├── fast_out_pve_1.nii.gz
├── fast_out_pve_2.nii.gz
├── fast_out_pveseg.nii.gz
├── fast_out_restore.nii.gz
├── fast_out_seg.nii.gz
├── mri_image.nii.gz
├── reoriented.nii.gz
├── roiimage.nii.gz
└── skull_stripped.nii.gz
Partial volume maps (pve) , for each class, where each voxel contains a value in the range 0–1 that represents the proportion of that class’s tissue present in that voxel.
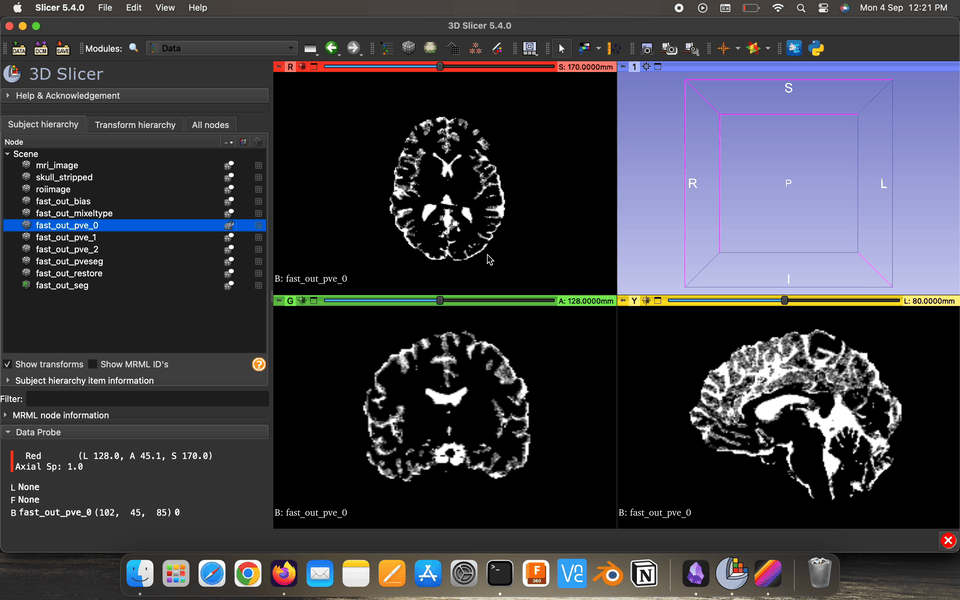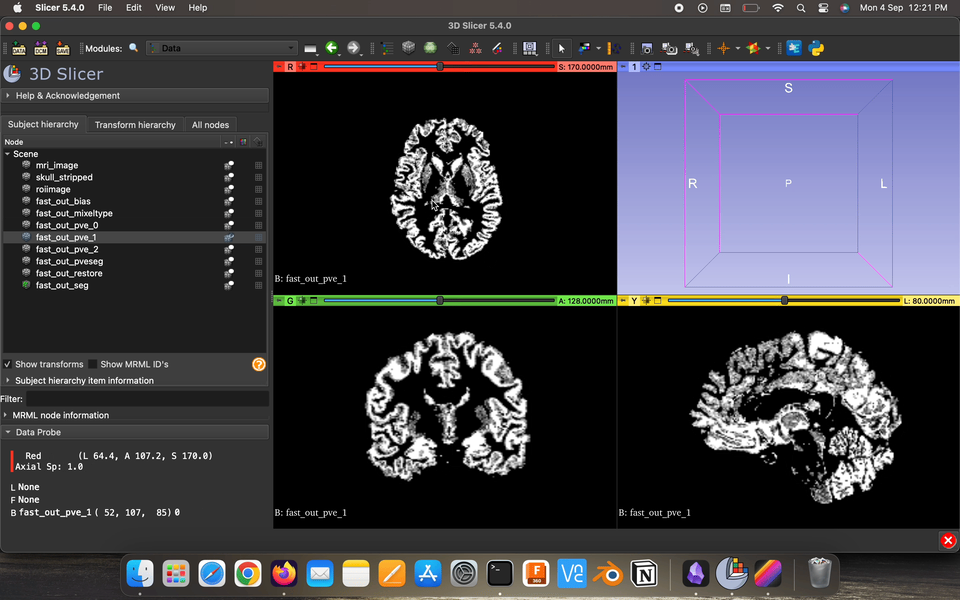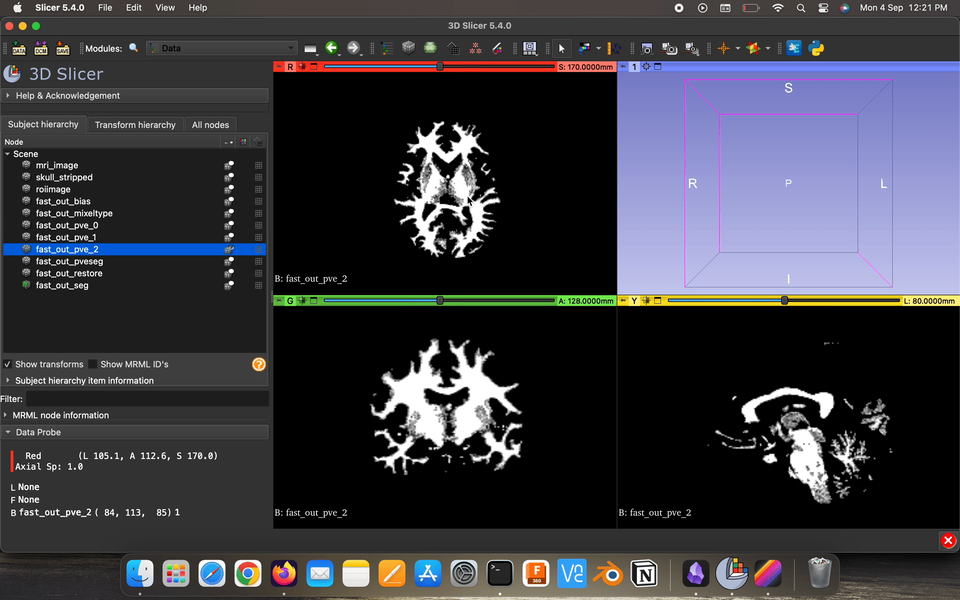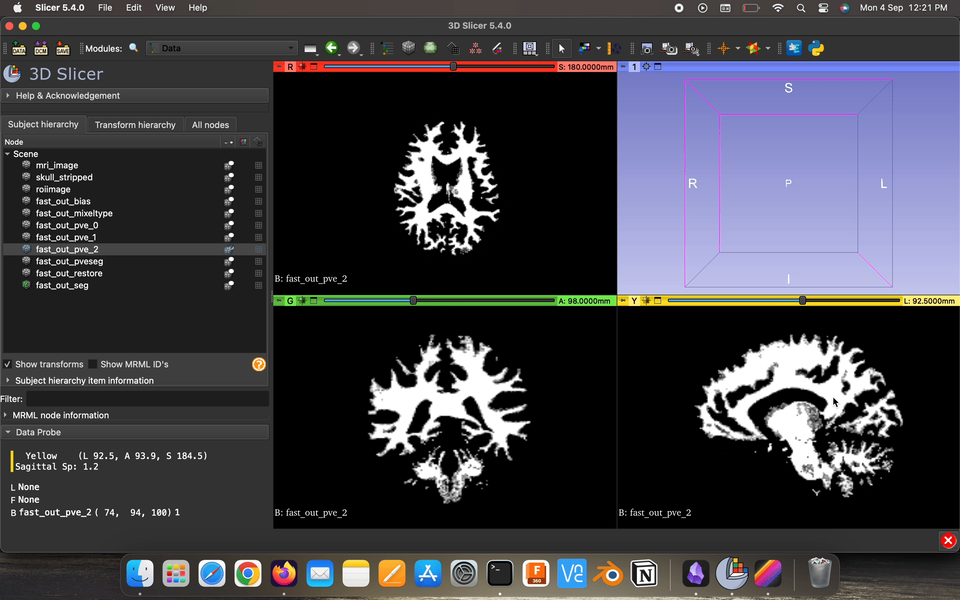
Restored input is the estimated restored input image after correction for bias field.

Bias field is the estimated bias field.

Fast Segmented images ,, has all the classes in a single MRI image

The brain segmentation is very useful in Volumetric caluclations, which we wont look into now,
We want the same cartesian / voxel co-ordinates to point us at the same anatomical structure ie, align the MRI image.
Why do we do so ?? Isn’t this tampering with the MRI data


Types of transformations


flirt -in fast_out_restore.nii.gz -ref /Users/ninad/fsl/data/standard/MNI152_T1_2mm.nii.gz -dof 12 -omat MRI_to_MNI.mat -out affine_trans
the above command does the affine transformation (12 DOF) to registers the MRI image to MNI158 standard 2mm template and furture help in the non linear transformation. -in the input MRI image path, -ref reference template (we used MNI 2mm template) -dof specifies DOF and i wanted Affine transformation so set it to 12, -omat outputs a 4x4 matrix in a .mat file with the affine transformation values, -out outputs the MRI image after registering to the reference image
cat MRI_to_MNI.mat
| 0.04992739769 -0.009461175045 1.376357676 -31.28157938 |
| -1.610147028 -0.1469847473 0.03976417704 252.6235512 |
| 0.182484106 -1.599149772 0.1794447228 206.7580262 |
| 0 0 0 1 |

The affine transformation can result in various changes like change in axis , for the visualization you can use the
fslreorient2std affine_trans.nii.gz reoriented_affine_tranform
There are many cost functions available in FLIRT, within-modality functions Least Squares and Normalised Correlation, as well as the between-modality functions Correlation Ratio (the default), Mutual Information and Normalised Mutual Information.
within-modality means the reference and the input MRI image should be of same type
fnirt --in=affine_trans.nii.gz --ref=/Users/ninad/fsl/data/standard/MNI152_T1_2mm -v
but it is not very likely it will do you any good. Fnirt has a large set of parameters that determine what is done, and how it is done. Without a knowledge of these parameters you will not get the best results that you can.
Efficiently using FNIRT
fnirt --in=roiimage.nii.gz --ref=/Users/ninad/fsl/data/standard/MNI152_T1_2mm.nii.gz --aff=MRI_to_MNI.mat --refmask=/Users/ninad/fsl/data/standard/MNI152_T1_2mm_brain_mask_dil.nii --config=/Users/ninad/fsl/etc/flirtsch/T1_2_MNI152_2mm.cnf --cout=fnirt_coef_warp --iout=fnirt_warped --fout=fnirt_warp --jout=fnirt_jacobian --refout=fnirt_ref_intensity --intout=fnirt_intensity_modulation -v --logout=fnirt_log

Is this what we were looking for ??
Comment below what went wrong…..? 🤔
Assignment : check out the config file located at fsl/etc/flirtsch/T1_2_MNI152_2mm.cnf and compare it with the options defined for the fnirt tool using — help option
Future work : Try to run fsl using R programming language using any fsl wrapper packages
Notes : always use -v (verbose) option in your commands to know what’s happening
References : FSL Course, Random code, FSL user guide